Write-Audit-Publish Pattern with Apache Iceberg on AWS using WAP id

This blog will cover how to implement a Write-Audit-Publish (WAP) Data Quality Pattern in AWS using Apache Spark on EMR that uses AWS Glue Catalog as metastore for the Apache Iceberg table and AWS Athena for querying the table.
This blog uses EMR 6.11, comes with Apache Spark 3.3 and, supports Apache Iceberg version 1.2.0
What is Write-Audit-Publish?
Write-Audit-Publish (WAP) is a data quality pattern commonly used in the data engineering workflow that helps us validate datasets by allowing us to write data to a staged snapshot/branch of the production table and fix any issues before finally committing the data to the production tables.
WAP using wap.id in action
Things to know before implementation
- On setting
write.wap.enabled=truein the Iceberg table property, all the data written in the table is instagemode only i.e. written data won't be reflected in the table until published. - Iceberg tables write data into
snapshots. - Snapshots in an Iceberg table can be seen by querying the snapshot metadata table.
- Snapshot metadata table can be queried via
<table-name>.snapshotswithinspark.sqlorSELECT * FROM "dbname"."tablename$snapshots"via Athena. - Once
write.wap.enabled=trueis set on the table, Athena doesn't show the table DDL until it'sUNSETagain but you can still query the data as usual. More details on this are covered here.
Pre-requisites for following along
An EMR running with Iceberg Cluster along with Spark 3.3 and Iceberg version < 1.2.1
The data that I will be using in this blog post is NYC Green Taxi Trip Data
When you are creating an EMR Cluster make sure, you Edit Software Setting > Enter Configuration
[{
"Classification":"iceberg-defaults",
"Properties":{"iceberg.enabled":"true"}
}]Software Configuration while Creating EMR
Preparing Table and Data for WAP
Let's start with setting up all the configurations that we need in SparkSession for this implementation to work.
%%configure
{
"conf": {
"spark.jars.packages": "com.amazon.deequ:deequ:2.0.3-spark-3.3",
"spark.jars.excludes": "net.sourceforge.f2j:arpack_combined_all",
"spark.sql.extensions": "org.apache.iceberg.spark.extensions.IcebergSparkSessionExtensions",
"spark.sql.catalog.glue": "org.apache.iceberg.spark.SparkCatalog",
"spark.sql.catalog.glue.catalog-impl":"org.apache.iceberg.aws.glue.GlueCatalog",
"spark.sql.catalog.glue.io-impl":"org.apache.iceberg.aws.s3.S3FileIO",
"spark.sql.catalog.glue.warehouse":"s3://bucket-name/glue/warehouse"
}
}SparkSession Configuration on Jupyter Notebook.
In the case of a spark script, you can set these configurations while creating yourSparkSessionusingSparkSession.builder.config("config-key", "config-value")orspark.conf.set("config-key", "config-value")
spark.jars.packagesandspark.jars.excludesare not required for WAP implementation with Iceberg. I have added these jars that will be used to demonstrate Auditing with Pydeequ.
To understand all the Iceberg related configurations, check here
Let's create an Iceberg Table that we will consider as a main Production Table on which we will implement WAP to write some new data from the Source Table.
from pyspark.sql.functions import lit, col
# Reading NYC Green Taxi Data for Sep 2023
raw_data_path = "s3://my-data-bucket/raw_data/nyc_tlc"
green_df = spark.read.format("parquet").load(f"{raw_data_path}/green/sep2023/")
# adding year and month column
green_df = green_df.withColumn("month", lit(9)) \
.withColumn("year", lit(2023))Reading Sep 2023 Green Taxi Trip Data for creating Production Iceberg Table
# creating an Iceberg table in glue catalog within nyc_tlc database. default compression is taken as 'zstd'
prod_table = "glue.nyc_tlc.green_taxi_trips"
green_df.writeTo(prod_table).partitionedBy("year", "month") \
.using("iceberg") \
.tableProperty("format-version", "2") \
.tableProperty("write.parquet.compression-codec", "snappy") \
.create()Writing data to create a Production Table called green_taxi_trips
## Check data into prod table
spark.table(prod_table).groupBy("year", "month").count().show()Verifying data into Production Table
+----+-----+-----+
|year|month|count|
+----+-----+-----+
|2023| 9|65471|
+----+-----+-----+Output
# checking snapshots for the table
spark.sql(f"select * from {prod_table}.snapshots").show(truncate=False)checking table snapshots
+-----------------------+-------------------+---------+---------+--------------------------------------------------------------------------------------------------------------------------------------------+-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|committed_at |snapshot_id |parent_id|operation|manifest_list |summary |
+-----------------------+-------------------+---------+---------+--------------------------------------------------------------------------------------------------------------------------------------------+-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|2024-01-02 08:47:33.829|4002909337217387123|null |append |s3://<glue_db_path>/green_taxi_trips/metadata/snap-4002909337217387123-1-a0d0113f-ad2e-4edf-a7a5-3716ca6c897b.avro|{spark.app.id -> application_1704184584490_0001, added-data-files -> 1, added-records -> 65471, added-files-size -> 1514631, changed-partition-count -> 1, total-records -> 65471, total-files-size -> 1514631, total-data-files -> 1, total-delete-files -> 0, total-position-deletes -> 0, total-equality-deletes -> 0}|
+-----------------------+-------------------+---------+---------+--------------------------------------------------------------------------------------------------------------------------------------------+-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+Output
Athena only supports Iceberg Table v2 tables. This is something to keep in mind if you are using Athena as query engine.
show create table nyc_tlc.green_taxi_trips
-- Result
CREATE TABLE nyc_tlc.green_taxi_trips (
VendorID int,
lpep_pickup_datetime timestamp,
lpep_dropoff_datetime timestamp,
store_and_fwd_flag string,
RatecodeID bigint,
PULocationID int,
DOLocationID int,
passenger_count bigint,
trip_distance double,
fare_amount double,
extra double,
mta_tax double,
tip_amount double,
tolls_amount double,
ehail_fee double,
improvement_surcharge double,
total_amount double,
payment_type bigint,
trip_type bigint,
congestion_surcharge double,
month int,
year int)
PARTITIONED BY (`year`, `month`)
LOCATION 's3://<glue_db_path>/nyc_tlc/green_taxi_trips'
TBLPROPERTIES (
'table_type'='iceberg',
'write_compression'='snappy'
);Checking Prod Table Schema using Athena
Let's prepare data that will be written using WAP in green_taxi_trips table.
## Source data that will be written later in the table.
source_df = spark.read.parquet(f"{raw_data_path}/green/oct2023/")
source_df = source_df.withColumn("month", lit(10)) \
.withColumn("year", lit(2023))
source_df.groupBy("year", "month").count().show()Reading data that will serve as Source Table Data and will be written in green_taxi_trips
+----+-----+-----+
|year|month|count|
+----+-----+-----+
|2023| 10|66177|
+----+-----+-----+Output for source_df
Alrighty everything is set to start with the WAP implementation now i.e.
- Production Table:
glue.nyc_tlc.green_taxi_trips - Source Data: data present in
source_df
WAP Implementation
Further steps show how to implement different stages in WAP i.e. Write - Audit - Publish with a bit of explanation.
Write
This stage ensures that we can safely write data into a branch created from the prod table so it's in isolation from the current production workloads.
- Set
write.wap.enabled=truein table properties for letting Iceberg table handle write using WAP Pattern
# Set write.wap.enabled=true for table to write data into WAP Pattern.
spark.sql(f"ALTER TABLE {prod_table} SET TBLPROPERTIES ('write.wap.enabled'='true')")Setting write.wap.enabled=true for table to write data using the WAP pattern
- Setting
wap.idin SparkSession that will be used to identify the staged snapshot
# As Branching is not available in Iceberg < 1.2.1,
# generated a wap.id that will be used to identify the snapshot_id for new data written into table
import uuid
wap_id = uuid.uuid4().hex
# set wap.id in spark session to put it in the summary of snapshot
spark.conf.set("spark.wap.id", wap_id)
print(wap_id)Generating and setting wap.id in current SparkSession
- Writing Source Data into Production Table
# Appending data into Iceberg Prod Table
source_df.writeTo(prod_table).append()Writing Source data into Production Table
# checking snapshots for the table
spark.sql(f"select * from {prod_table}.snapshots").show(truncate=False)Checking table snapshots and wap.id in the snapshot summarysnapshots
+-----------------------+-------------------+-------------------+---------+--------------------------------------------------------------------------------------------------------------------------------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|committed_at |snapshot_id |parent_id |operation|manifest_list |summary |
+-----------------------+-------------------+-------------------+---------+--------------------------------------------------------------------------------------------------------------------------------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|2024-01-02 08:47:33.829|4002909337217387123|null |append |s3://<glue_db_path>/green_taxi_trips/metadata/snap-4002909337217387123-1-a0d0113f-ad2e-4edf-a7a5-3716ca6c897b.avro|{spark.app.id -> application_1704184584490_0001, added-data-files -> 1, added-records -> 65471, added-files-size -> 1514631, changed-partition-count -> 1, total-records -> 65471, total-files-size -> 1514631, total-data-files -> 1, total-delete-files -> 0, total-position-deletes -> 0, total-equality-deletes -> 0} |
|2024-01-02 08:56:07.165|7270839791374367070|4002909337217387123|append |s3://<glue_db_path>/green_taxi_trips/metadata/snap-7270839791374367070-1-26181e44-1f0d-47e2-8e88-febb7f2c1e7d.avro|{spark.app.id -> application_1704184584490_0001, wap.id -> fcd54f05ee4b4480a21c191cd5c17f39, added-data-files -> 1, added-records -> 66177, added-files-size -> 1519040, changed-partition-count -> 1, total-records -> 131648, total-files-size -> 3033671, total-data-files -> 2, total-delete-files -> 0, total-position-deletes -> 0, total-equality-deletes -> 0}|
+-----------------------+-------------------+-------------------+---------+--------------------------------------------------------------------------------------------------------------------------------------------+----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+Output. Check wap.id in summary column
Let's verify the data in the Production Table
spark.table(prod_table).groupBy("year","month").count().show()Verifying data in Production Table
+----+-----+-----+
|year|month|count|
+----+-----+-----+
|2023| 9|65471|
+----+-----+-----+Output from Production Table
As mentioned before, the data that is written with write.wap.enabled=true is in staging mode. Let's verify the data in the snapshot written in the previous write.
# Getting the snapshot_id corresponding to wap_id that was added in Spark Session.
from pyspark.sql.functions import col
staged_data_snap_id = spark.sql(f"select snapshot_id, summary['wap.id'] as wap_id from {prod_table}.snapshots").filter(col("wap_id") == wap_id).collect()[0]['snapshot_id']
# Reading data in the staged snapshot
staged_data = spark.read.option("snapshot-id", staged_data_snap_id).table(prod_table)
staged_data.groupBy("year","month").count().show()Checking data in the staged snapshot
+----+-----+-----+
|year|month|count|
+----+-----+-----+
|2023| 10|66177|
|2023| 9|65471|
+----+-----+-----+Output from staged_snapshot
Audit
This stage enables us to understand the quality of our newly inserted data. During this stage, we can run various data quality checks such as inspecting NULL values, identifying duplicated data, etc.
You can define any of the DQ Processes here via some code, third-party provider, or any other service. A few of such options are:
- via PyDeequ
- via AWS Glue Data Quality
- via Pyspark/Pandas etc
I will be using the first option here via PyDeequ. To learn more about PyDeequ and how it works, you can refer to this blog post.
Let's say our DQ requirement is:
- Completeness criteria for VendorID should be 1.0 i.e. there are no `null`.
- total_amount shouldn't be negative
- payment_type should be discrete numbers between 1 and 6.
import os
# Setting SPARK_VERSION in environment variable for Pydeequ
os.environ['SPARK_VERSION'] = '3.3'
from pydeequ.checks import *
from pydeequ.verification import *
# Check represents a list of constraints that can be applied to a provided Spark Dataframe
vendorID_checks = Check(spark, CheckLevel.Error, "VendorID Checks")
payment_type_checks = Check(spark, CheckLevel.Error, "payment_type Checks")
tot_amt_checks = Check(spark, CheckLevel.Warning, "total_amount Checks")
checkResult = VerificationSuite(spark) \
.onData(staged_data) \
.addCheck(vendorID_checks.isComplete("VendorID")) \
.addCheck(payment_type_checks.isContainedIn("payment_type", ['1','2','3','4','5','6'])) \
.addCheck(tot_amt_checks.isNonNegative("total_amount")) \
.run()
print(f"Verification Run Status: {checkResult.status}")
checkResult_df = VerificationResult.checkResultsAsDataFrame(spark, checkResult)
checkResult_df.show(truncate=False)Checking Data Quality using PyDeequ
+-------------------+-----------+------------+--------------------------------------------------------------------------------------------------------------------------------------------------+-----------------+-------------------------------------------------------------------+
|check |check_level|check_status|constraint |constraint_status|constraint_message |
+-------------------+-----------+------------+--------------------------------------------------------------------------------------------------------------------------------------------------+-----------------+-------------------------------------------------------------------+
|VendorID Checks |Error |Success |CompletenessConstraint(Completeness(VendorID,None)) |Success | |
|payment_type Checks|Error |Success |ComplianceConstraint(Compliance(payment_type contained in 1,2,3,4,5,6,`payment_type` IS NULL OR `payment_type` IN ('1','2','3','4','5','6'),None))|Success | |
|total_amount Checks|Warning |Warning |ComplianceConstraint(Compliance(total_amount is non-negative,COALESCE(CAST(total_amount AS DECIMAL(20,10)), 0.0) >= 0,None)) |Failure |Value: 0.9968780384054449 does not meet the constraint requirement!|
+-------------------+-----------+------------+--------------------------------------------------------------------------------------------------------------------------------------------------+-----------------+-------------------------------------------------------------------+PyDeequ Data Quality Results
As we can see, one of the DQ Validations failed as total_amount column has some negative values.
Based on this result and discussion with stakeholders, we can decide whether we want to Publish the data to the Production table or not.
Scenario 1: Let's say as per discussion with stakeholders, we don't want to publish the data in Production because DQ Checks have failed.
spark.sql(f"CALL glue.system.expire_snapshots(table => '{prod_table}', snapshot_ids => ARRAY({staged_data_snap_id}))")Rejecting/Expiring the snapshot based on DQ results
Scenario 2: Let's say as per discussion with stakeholders, we can clean the data by filtering out all such records that have negative total_amount and write the data with a negative total_amount to some bad data location.
# filtering out the non-negative data from staged_df
cleaned_df = staged_data.filter(col("total_amount") >= 0)
# writing bad data into an S3 location that can be looked on later.
bad_data_df = staged_data.filter(col("total_amount") < 0)
bad_data_df.write.partitionedBy("year", "month").parquet("bad-data-s3-location")Filtering out records having total_amount < 0 to make it clean
Once we have cleaned the data, we can rewrite this into the table.
# write cleaned data into prod table.
cleaned_df.writeTo(prod_table).overwritePartitions()Writing cleaned data into table.
Now the important thing to notice here is that as the data is re-written on top ofstagedsnapshot, a newsnapshot_idfor this write is generated . This newsnapshot_idwill be the actual data, that needs to be published in the Production Table.
Let's look into the snapshot metadata table to understand what I mean.
# Checking the snapshots again
spark.sql(f"select * from {prod_table}.snapshots").show(truncate=False)Checking snapshot_ids corresponding to the wap_id
+-----------------------+-------------------+-------------------+---------+--------------------------------------------------------------------------------------------------------------------------------------------+-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|committed_at |snapshot_id |parent_id |operation|manifest_list |summary |
+-----------------------+-------------------+-------------------+---------+--------------------------------------------------------------------------------------------------------------------------------------------+-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|2024-01-02 08:47:33.829|4002909337217387123|null |append |s3://<glue_db_location>/green_taxi_trips/metadata/snap-4002909337217387123-1-a0d0113f-ad2e-4edf-a7a5-3716ca6c897b.avro|{spark.app.id -> application_1704184584490_0001, added-data-files -> 1, added-records -> 65471, added-files-size -> 1514631, changed-partition-count -> 1, total-records -> 65471, total-files-size -> 1514631, total-data-files -> 1, total-delete-files -> 0, total-position-deletes -> 0, total-equality-deletes -> 0} |
|2024-01-02 08:56:07.165|7270839791374367070|4002909337217387123|append |s3://<glue_db_location>/green_taxi_trips/metadata/snap-7270839791374367070-1-26181e44-1f0d-47e2-8e88-febb7f2c1e7d.avro|{spark.app.id -> application_1704184584490_0001, wap.id -> fcd54f05ee4b4480a21c191cd5c17f39, added-data-files -> 1, added-records -> 66177, added-files-size -> 1519040, changed-partition-count -> 1, total-records -> 131648, total-files-size -> 3033671, total-data-files -> 2, total-delete-files -> 0, total-position-deletes -> 0, total-equality-deletes -> 0} |
|2024-01-02 09:14:03.382|8254249659682978316|4002909337217387123|overwrite|s3://<glue_db_location>/green_taxi_trips/metadata/snap-8254249659682978316-1-70060bcd-8b81-4e00-b26c-92ecca1145bd.avro|{spark.app.id -> application_1704184584490_0001, replace-partitions -> true, wap.id -> fcd54f05ee4b4480a21c191cd5c17f39, added-data-files -> 2, deleted-data-files -> 1, added-records -> 131237, deleted-records -> 65471, added-files-size -> 3022251, removed-files-size -> 1514631, changed-partition-count -> 2, total-records -> 131237, total-files-size -> 3022251, total-data-files -> 2, total-delete-files -> 0, total-position-deletes -> 0, total-equality-deletes -> 0}|
+-----------------------+-------------------+-------------------+---------+--------------------------------------------------------------------------------------------------------------------------------------------+-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+Output from snapshot metadata table
Things to observe in above output:
- Now we have 2
snapshot_id(7270839791374367070and8254249659682978316) corresponding to thewap.idi.e.fcd54f05ee4b4480a21c191cd5c17f39written at different times i.e.commited_at - Data that needs to be published in the Production Table should be the cleaned data i.e. of
snapshot_id=8254249659682978316
Publish
This stage allows data to be made available to the production tables for consumption by the downstream analytical applications.
Once the Auditing is done and DQ is as expected or fixed. We can Publish the final snapshot in the Production table by calling an Iceberg Store Procedure called system.cherrypick_snapshot.
To publish the snapshot from the above scenario, we need to get the latest snapshot_id based on commited_at timestamp for the wap.id that we used to perform the write.
# getting the latest snapshot_id
udpated_data_snap_id = spark.sql(f"select snapshot_id, summary['wap.id'] as wap_id from {prod_table}.snapshots").filter(col("wap_id") == wap_id).orderBy(col("committed_at").desc()).collect()[0]['snapshot_id']
# Publishing the this new snapshot_id into Production table
spark.sql(f"CALL glue.system.cherrypick_snapshot('{prod_table}', {udpated_data_snap_id})").show()Getting latest snapshot_id and publishing it into the Production Table
+-------------------+-------------------+
| source_snapshot_id|current_snapshot_id|
+-------------------+-------------------+
|8254249659682978316|8254249659682978316|
+-------------------+-------------------+Output after running cherrypick_snapshot to Publish data into the Production Table
Let's verify the records in the Production Table now from AWS Athena
select "year", "month", count(*) from nyc_tlc.green_trips_data group by 1,2Validating the final data in Athena Table
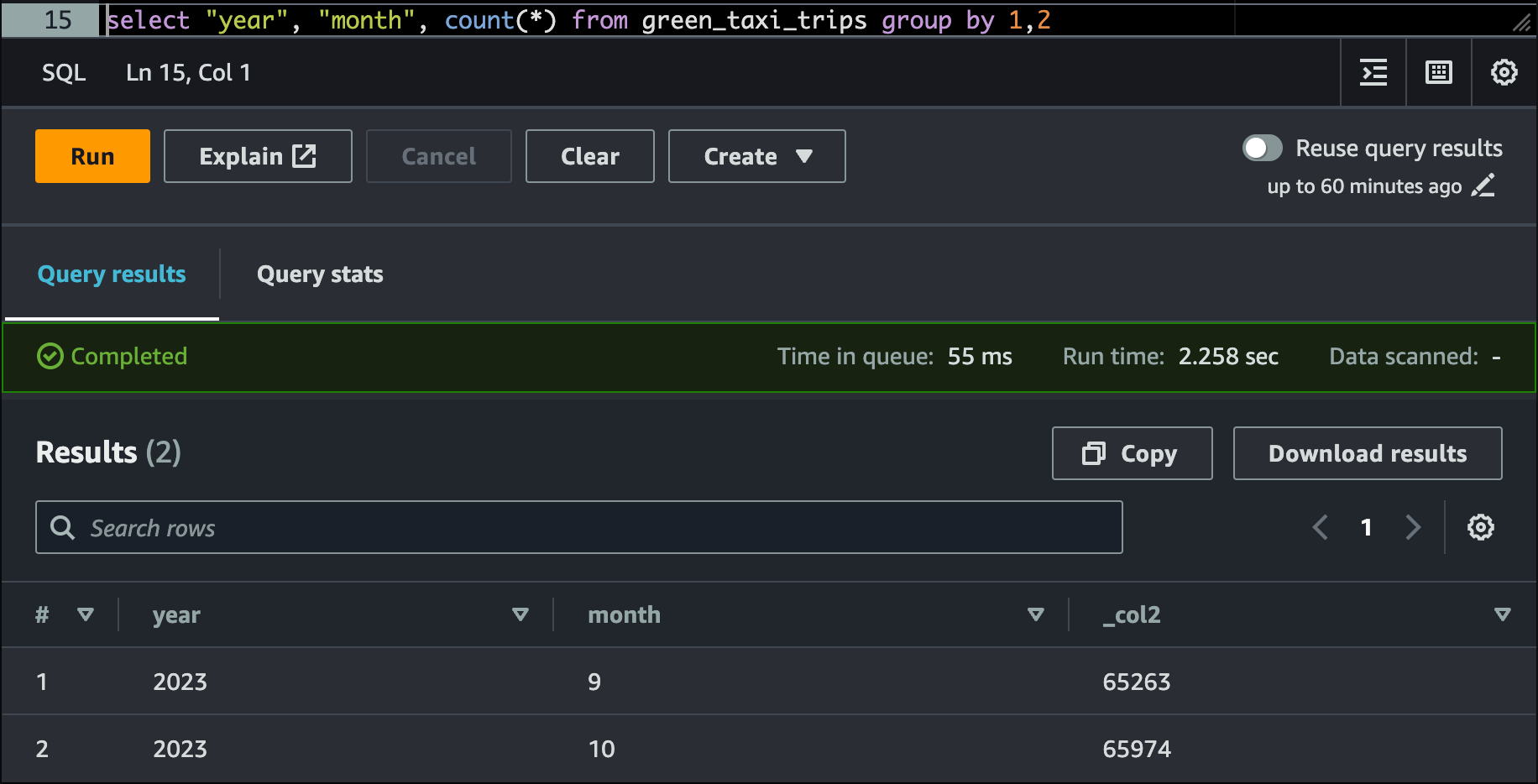
Finally, to make the DDL queryable again from Athena, UNSET the table property write.wap.enabled . The reason for doing this is explained in detail in this blog post.
# unset from table properties so that create table ddl can be seen again on Athena.
spark.sql(f"ALTER TABLE {prod_table} UNSET TBLPROPERTIES ('write.wap.enabled')")Unsetting write.wap.enabled from the Iceberg Table
That's it for this one folks..!! Now you know how to implement WAP Pattern on AWS for Iceberg version < 1.2.1
If you have read until here, please leave a comment below, and any feedback is highly appreciated. See you in the next post..!!! 😊
If it has added any value to you and want to read more content like this, subscribe to the newsletter, it's free of cost and I will make sure every post is worth your time.





Member discussion